MRI-based advanced imaging analysis and machine learning may help predict "molecular profiles" in patients with hepatocellular carcinoma (HCC), according to research published August 5 in Radiology.
The finding is from a proof-of-concept study in 75 patients who underwent T1-weighted, contrast-enhanced, dynamic MRI prior to treatment and opens new avenues for personalizing care, noted lead author Nickolai Matuschewski, MD, of Yale University in New Haven, CT, and colleagues.
"Given that oncogenic pathways drive tumor growth and vascularization patterns, MRI could be used for whole-tumor molecular profiling," the group wrote.
Currently, biopsies are the only diagnostic tool that can provide direct pretreatment tissue characterization, important information for determining treatment approaches. However, biopsies can be limited because of sampling bias, which is a substantial concern in patients with HCC due to intratumoral heterogeneity, the authors explained.
In contrast, MRI can provide an assessment of whole-tumor morphology with additional characterization of the surrounding liver. Moreover, modeling correlations between imaging and molecular characterization has emerged as a promising strategy for comprehensive pretreatment molecular profiling, the group noted.
To provide further evidence to support the value of the technique, the researchers first gathered data on 75 treatment-naive patients with HCC who underwent MRI prior to tumor resection or liver transplantation. Next, they used software (PyRadiomics v3.0.1) to extract radiomic imaging features. From the original images, they identified 19 first-order statistics and 13 shape-based and 75 texture features per volume of interest.
Meanwhile, pathology analysis of the resected HCC tumor samples was performed via immunohistochemistry to assess p53 loss of function, CTNNB1 activation, FoXM1 activation, and PD-L1, pAKT, pSMAD2/3, and SOAT1 expression. These are markers that provide insight into the biology and behavior of tumors.
Finally, the researchers used machine learning to analyze clinical and imaging features and their association with HCC molecular profiling.
 A graphic abstract of the study.RSNA
A graphic abstract of the study.RSNA
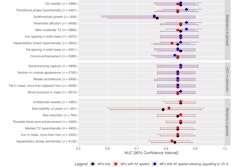
The MRI data predicted p53 and CTNNB1 activation with areas under the receiver operating characteristic curve (AUCs) of 0.79 and 0.7.
Quantitative imaging data also predicted the immuno-oncological profile of PD-L1 expression (AUC = 0.85).
Quantitative imaging data predicted CTNNB1 status with a sensitivity of 83% and specificity of 61%.
"Quantitative MRI markers derived from whole tumor and liver volumes can potentially predict p53, PD-L1, and CTNNB1 status in HCC," the group wrote.
Ultimately, this was a proof-of-concept study that warrants more research, yet the study highlights that imaging-based screening with targeted pathological workup could help refine treatment strategies, the researchers concluded.
In an accompanying editorial, Roberto Cannella, MD, of the University of Palermo in Italy, noted that identifying HCC subtypes with CTNNB1 mutations using quantitative MRI could be particularly important, as the mutation is seen in about 40% of patients and is associated with distinct prognostic and therapeutic implications.
"CTNNB1 mutations have been associated with lack of immune infiltration in the tumors, which has important therapeutic implications, including a limited response to immunotherapy treatments," he wrote.
Ultimately, the findings of the study could serve as a foundation for future investigations aimed at delivering a comprehensive assessment of HCC to guide personalized treatment strategies, Cannella concluded.
The full study is available here.