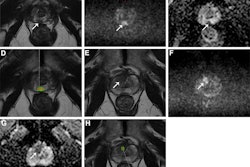
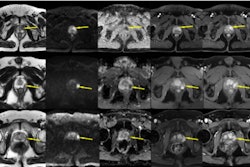
PI-RADS v2.1 shows high sensitivity for categorizing clinically significant prostate cancer on MRI at both the patient and lesion levels, according to a literature review published October 15 in the American Journal of Roentgenology.
A group led by Andrea Nedelcu, MD, of the University of Freiburg in Germany did find, however, that several studies included in the review had a high risk of bias or applicability concerns -- and were thus associated with reduced sensitivity for PI-RADS category 2 (i.e., clinically significant cancer unlikely).
"[Our] results confirm overall high sensitivity of PI-RADS v2.1 for clinically significant prostate cancer detection, supporting the system's use for prostate MRI interpretation," the team wrote. "However, a considerable proportion (29%) of studies had a high risk of bias and/or high concerns of applicability … suggesting potentially flawed estimates resulting from inclusion of investigations with quality issues."
PI-RADS v2.1 was jointly developed by the American College of Radiology (ACR), the European Society of Urogenital Radiology (ESUR), and the Admetech Foundation and released in 2019. Its goal is to "help improve early diagnosis of clinically significant prostate cancer and reduce unnecessary biopsies and treatment for benign and subclinical diseases," according to the ACR.
But "estimates of outcome metrics for PI-RADS version 2.1 have shown substantial heterogeneity, possibly relating to risks of bias in the relevant literature," the study authors explained. To assess the quality of the literature on this topic, they searched seven databases and registers for research published between March 2019 and September 2023 that reported diagnostic test accuracy metrics and/or cancer detection rates of PI-RADS v2.1 for identifying clinically significant prostate cancer in men suspected to have the disease.
The analysis included 117 studies with 25,228 patients and 15,553 lesions. The group used the QUADAS-2 tool to rate the studies' risk of bias and concerns of applicability. (QUADAS-2 assesses the quality of studies by evaluating research domains such as patient selection, the new procedure or protocol being investigated and its reference standard, and how and when tests/protocols were administered to study participants). It also calculated estimates of sensitivity and specificity of PI-RADS categories.
In all the studies, the investigators rated at least one of the QUADAS-2 domains as unclear or of high risk of bias or concerns of applicability. They also noted that 29% of the studies had a high risk of bias or concerns of applicability overall.
But PI-RADS v2.1 did show high sensitivity and specificity for categorizing patient and lesion-level prostate cancer risk:
PI-RADS performance for categorizing prostate cancer risk | ||
Measure | PI-RADS category ≥3 (intermediate risk of disease) | PI-RADS category ≥4 (high risk of disease) |
| Patient-level sensitivity | 96% | 88% |
| Patient-level specificity | 43% | 66% |
| Lesion-level sensitivity | 96% | 89% |
| Lesion-level specificity | 44% | 63% |
The investigators also found that lesion-level sensitivity for PI-RADS category ≥4 was lower for high-risk studies than for remaining studies (78% vs. 89%; p = 0.008).
Finally, the group reported the following:
Patient-level cancer detection rate by PI-RADS category | |||||
Measure | PI-RADS category 1 | PI-RADS category 2 | PI-RADS category 3 | PI-RADS category 4 | PI-RADS category 5 |
| Cancer detection rate | 3% | 6% | 20% | 53% | 83% |
Nedelcu and colleagues noted that their review did confirm PI-RADS v2.1's high sensitivity and specificity for categorizing prostate cancer risk, but underscored that "heterogeneity in reported study results remained a substantial issue for specificity and cancer detection rates." They also wrote that "additional factors beyond study quality appear to be key drivers of [the mixed results], and concluded that "these observations should guide ongoing research regarding PI-RADS v2.1 performance, to increase the quality of future evidence and raise awareness of issues impacting the translation of study findings into routine clinical practice."
The complete study can be found here.