Texas researchers have developed a new knowledge-based segmentation technique that surpasses previous efforts to segment the brain accurately with MRI, according to a new report in the Journal of Medical Imaging.
The new approach is better equipped to handle a difficult task in segmentation -- managing signal intensity overlap between tissues -- than previous methods, the authors wrote. It should eventually lead to better identification of boundaries between tissues, lead author Nishant Verma, PhD, from the University of Texas at Austin, told AuntMinnie.com.
 Nishant Verma, PhD, from the University of Texas at Austin.
Nishant Verma, PhD, from the University of Texas at Austin."Ultimately, we are trying to solve the problem that there can be much more effective classification at the boundary" between tissue types, he said.
The strategy of using information from real clinical data, in the form of adaptive tissue class priors from imaging atlases, produced "a more accurate classification of voxels belonging to intensity overlap regions in comparison with the existing methods, several of which employed methods for correction of image corruptions," Verma and colleagues wrote (J Med Imaging, October 2014, Vol. 1:3).
Segmentation is the 1st step
MR tissue segmentation is an important component of any comprehensive MR image analysis. It involves categorizing brain MR voxels into four classes: white matter, gray matter, cerebrospinal fluid, and background.
But in MRI the task is complicated by the presence of image corruption, including partial-volume effects and intensity homogeneities. Segmenting the brain requires incorporating the contributions from the corruptions along with the rest of the image data while categorizing MR voxels into the four classes, Verma and colleagues Gautam Muralidhar, Alan Bovik, Matthew Cowperthwaite, Mark Burnett, and Mia Markey wrote.
"Most errors in segmentation are actually on the boundaries between tissues, where you see a combination of different tissues," Verma said. "Also, there are major artifacts on MRI that may further worsen this problem, so even more confusion happens on these boundaries."
Verma's group developed a new knowledge-driven decision theory (KDT) approach, which models assumptions about intensity distributions within the tissue classes using a nonparametric method known as kernel density estimation. The knowledge about overlapping tissue intensities comes from a combination of probabilistic atlas maps and spatial and contextual information obtained from Markov random fields to guide volumetric MR tissue segmentation.
"We looked at the overlap of tissues and plotted it," Verma said.
KDT actually embeds the known tissue overlap trends into the algorithm to correct for partial-volume artifacts; it then superimposes them with Markov random fields in subsequent iterations. The idea of the study was to distinguish, for example, gray matter and cerebrospinal fluid without interference from preprocessing techniques, Verma said.
Incorporating intensity overlap knowledge in the segmentation model permits more accurate classification of voxels that reside in the intensity overlap spectrum, the authors wrote. In KDT, a decision theory-based objective function is minimized using a variational level set-based approach.
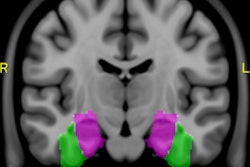
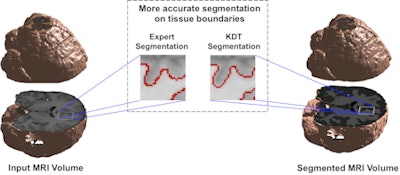 Brain segmentation is improved with use of the KDT method. Image courtesy of Nishant Verma, PhD.
Brain segmentation is improved with use of the KDT method. Image courtesy of Nishant Verma, PhD.2 types of MR data
The researchers evaluated the proposed method on two well-established real MR datasets with previously performed expert manual segmentations, and then compared the results with existing segmentation algorithms.
The group obtained the two MR datasets from the Internet Brain Segmentation Repository (IBSR). The first dataset (IBSR-20) contained MR volumes from 20 normal subjects along with ground-truth tissue segmentations performed by experts. The data were acquired on a 1.5-tesla scanner with a slice thickness of 1.5 mm. These samples were chosen because they present varying levels of segmentation difficulty, such as low-contrast and high-intensity inhomogeneity.
The second dataset (IBSR-18) included MR volumes from 18 normal subjects under IBSR V2.0. These data have higher spatial resolution than the first dataset and were collected using a 3-tesla scanner at a slice thickness of 1.5 mm. All MR volumes in both datasets underwent automatic skull stripping using the brain extraction tool as a preprocessing step.
For the prior knowledge, or adaptive tissue class priors, the researchers used the International Consortium for Brain Mapping atlas maps provided by the Laboratory of Neuroimaging at the University of California, Los Angeles. Aligning atlas maps with subject MR data from 18 subjects was performed using the Oxford Centre for Functional Magnetic Resonance Imaging of the Brain's (FMRIB) Linear Image Registration Tool (FLIRT).
Better segmentation results
KDT produced statistically significant improvements in the segmentation accuracies of white matter and grey matter over the next most accurate method described by Rivera, meaning that KDT has better white-matter segmentation performance than the other methods, the researchers found. Three of these have similar white-matter segmentation results, but KDT yielded better average gray-matter and cerebrospinal fluid segmentation accuracies.
The summary segmentation accuracies were 79.72 ± 2.78 for white matter, 88.59 ± 1.23 for gray matter, and 74.39 ± 6.43 for cerebrospinal fluid when the three MR volumes used for parameter optimization were removed, suggesting that there is no significant bias of the chosen parameter values on the segmentation results, the authors wrote.
Not surprisingly, in comparison with IBSR-20, KDT produced better segmentation accuracies for IBSR-18 MR volumes due to the higher resolution of MR volumes in the IBSR-18 dataset acquired at 3 tesla, they wrote.
KDT outperforms most existing segmentation methods for simultaneously segmenting brain MR images into white matter, gray matter, and cerebrospinal fluid, they wrote. Some previous methods have reported similar segmentation performance on certain tissue types, but they don't perform as well on other tissue types.
"KDT also performs better than the popular segmentation method [FMRIB's Automated Segmentation Tool (FAST)], which is widely used by the neuroimaging community," they wrote.
Still, while KDT does a better job of handling intensity overlaps between tissue classes, it is affected by high levels of intensity homogeneities, the authors wrote. In the study, segmentation performance of KDT declined in MRI volumes with high levels of intensity inhomogeneities. The use of preprocessing steps can reduce these homogeneities, but it increases the overall computational complexity and was rejected for KDT. As predicted, with KDT the computational complexity was reduced.
"When we compared this complexity with other papers which did an analysis, we saw that ours is much lower than those methods," Verma said. "Another way to compare complexity is that we compared the physical run times on our machine," and again KDT came out on top.
KDT took four minutes to segment an image with 60 slices in a volume and nine minutes for an image with 185 slices, he said. This compares to six minutes and 10 minutes of computing time, respectively, using the FAST method.
"Segmentation is the first step in most clinical decision-support systems, so it's an application area where a lot of people are working," Verma said.
It's the first step for Parkinson's disease, Alzheimer's disease, and others, for which segmentation results are used to develop biomarkers.
"Most of the information is in the form of white matter or gray matter or even the subcortical structure" of the brain, he said. "If you use that, you can predict if the patient will get Alzheimer's, so segmentation is one of the most essential steps, and definitely accuracy in segmentation would improve the predictive power of these decision-support systems."
After refining the code, the team plans to make it available in the University of Texas Biomedical Informatics Lab, Verma said.