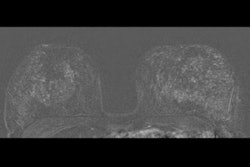
A radiomics model developed by an Italian research team shows promise in differentiating malignant from benign breast lesions on dynamic contrast-enhanced MR images, according to a study published September 29 in Academic Radiology.
The team, led by Carmelo Militello, PhD, from the Italian National Research Council, touted the high accuracy and specificity of their predictive model, which could help avoid unnecessary biopsies.
"Considering that we analyzed only the strongest dynamic contrast-enhanced-MRI phase, thus enabling an efficient and less protocol-dependent approach ... our results are consistent with recent literature studies," Militello and colleagues wrote.
Diagnosing early invasive breast cancer needs clinical evaluation, radiological imaging, and image-guided biopsy. MRI is well-suited for local staging of breast cancer due to its high sensitivity. However, it has limited specificity when it comes to characterizing lesions as malignant or benign, which could lead to unnecessary additional imaging or biopsy.
Dynamic contrast-enhanced MRI has shown promise in previous studies by assessing tumor vascularity and defining enhancement kinetic curves with high sensitivity. But its specificity is still on the lower end.
Militello et al wanted to develop a feasible, radiomics-powered model to be integrated into the clinical practice by exploiting standard-of-care MRI and reducing the required image preprocessing.
They looked at 107 radiomic features extracted from a dataset of 111 patients (110 women, 1 man) in a single setting. The average age of the patients was 47.83 years. Out of the total number, 41 patients had lesions classified as BI-RADS 3, 15 as BI-RADS 4, and 55 as BI-RADS 5.
A preprocessing step was performed for the model to find only robust nonredundant features. A support vector machine (SVM) was trained in a nested five-fold cross-validation scheme by exploiting several unsupervised feature selection methods. The support vector machine was combined with the following features: unsupervised discriminative feature selection (UDSF), dependence guided unsupervised feature selection (DGUFS), and unsupervised feature selection with ordinal locality (UFSOL).
The predictive model performance was evaluated in terms of area under the receiver operating characteristic (AUROC), specificity, sensitivity, positive predictive value, and negative predictive value.
| Performance of radiomics model for characterizing lesions on breast MRI | |||
| DGUFS + SVM | UFSOL + SVM | UDFS + SVM | |
| AUROC | 0.723 | 0.708 | 0.725 |
| Specificity | 0.741 | 0.62 | 0.741 |
| Sensitivity | 0.705 | 0.796 | 0.709 |
| Positive predictive value | 0.719 | 0.669 | 0.72 |
| Negative predictive value | 0.747 | 0.777 | 0.75 |
Despite different combinations showing advantages in certain measurement areas, the UDFS + SVM combination achieved the best average performance on the blinded test set.
The researchers said combining radiomics features extracted from different types of sequences could increase their model's predictive power.
They also said they are planning to extend their study to a larger patient cohort and to nonmass enhancement breast lesions, saying a multicentric study would allow for assessing the generalization abilities of the developed model. The team highlighted the need for prospective studies to explore how such models could be deployed into the clinical practice to characterize breast masses in dynamic contrast-enhanced MRI.
"Complementing studies on BI-RADS 4 and 5 characterization, analyzing BI-RADS 3 lesions by an ad-hoc method would be clinically relevant to avoid unnecessary biopsies and resulting in an optimization of the current patient management," the study authors wrote.