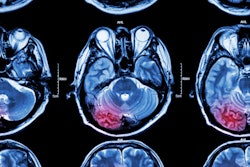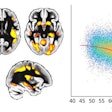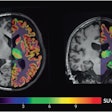
An artificial intelligence (AI) algorithm can predict -- based on baseline MRI and prior to intervention -- the final infarct lesions in patients with acute ischemic stroke, according to research published online March 12 in JAMA Network Open. It can even outperform current clinical prediction methods.
A team of researchers from Stanford University reported that their deep-learning algorithm yielded strong predictive performance for subacute infarction 3 to 7 days old.
"Predicting the subacute infarct lesion may help clinicians prepare for decompression treatment and aid in patient selection for neuroprotective clinical trials," wrote the authors, led by first author Dr. Yannan Yu and senior author Dr. Greg Zaharchuk, PhD.
The ability to predict the size and location of an infarct is important for clinical decision-making and prognosis in patients with acute ischemic stroke. Utilizing the initial MRI exams as inputs for the model, the researchers trained a U-net -- a type of deep convolutional neural network -- to predict the final infarct lesions.
Training, validation, and testing of the algorithm was performed using patients from the Imaging Collaterals in Acute Stroke (iCAS) study from April 14-15, 2018 and the Diffusion and Perfusion Imaging Evaluation for Understanding Stroke Evolution-2 (DEFUSE-2) study from July 14, 2008 to September 17, 2011.
The patients were grouped based on 24-hour imaging results into four categories: unknown, minimal, partial, and major reperfusion. The final true infarct lesions at 3 to 7 days was considered the ground truth for the model.
The algorithm calculates the probability of infarction for every voxel in the image, and these results are then processed to produce a final prediction, according to the researchers. In the 182 patients included in the model, the model yielded a median area under the curve of 0.93 and a dice score coefficient (DSC) of 0.53; a DSC score of 0.5 or greater indicates significant overlap, according to the authors. The model also had a median volume error of 9 mL.
The researchers also compared the algorithm's performance with two prediction methods that are used clinically to select patients for endovascular therapy: the apparent diffusion coefficient (ADC) of the ischemic core and the perfusion parameter time to maximum of the residue function (Tmax) of the penumbra.
In a subgroup of patients with minimal reperfusion for which comparison with existing clinical methods was possible, the deep-learning model produced comparable performance (p = 0.37). However, the algorithm outperformed the clinical methods in a subgroup of patients with major reperfusion (p = 0.002).
"The model was trained without including information about reperfusion status, yet it had comparable performance in patients with and without major reperfusion compared with a common clinically used ADC and Tmax thresholding software package," the authors wrote. "Furthermore, it performed similarly well in patients with partial or unknown reperfusion status, among whom neither of the traditional prediction methods based on the diffusion-perfusion mismatch paradigm apply."
Further studies are now warranted in larger and more diverse patient cohorts and to improve the model's performance by incorporating important clinical predictors, according to the researchers.