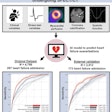
Researchers have developed an artificial intelligence (AI) deep-learning algorithm that may help clinicians diagnose parkinsonian diseases, according to research published in the April issue of the Journal of Nuclear Medicine.
Chinese researchers from Fudan University in Shanghai used a 3D deep convolutional neural network to extract deep metabolic imaging indices from F-18 FDG-PET scans. The model effectively differentiated between Parkinson's disease and other parkinsonian syndromes, such as multiple system atrophy and progressive supranuclear palsy, according to findings.
"This work confirms that the emerging artificial intelligence can extract in-depth information from molecular imaging to enhance the differentiation of complex physiology," said lead author Dr. Ping Wu, PhD, in a news release from the Society for Nuclear Medicine and Molecular Imaging (SNMMI).
More than 10 million people worldwide live with Parkinson's disease. Accurate diagnosis of the disease is often a challenge, particularly in the early stages, as its symptoms overlap considerably with those of other atypical parkinsonian syndromes.
In this study, the researchers built a 3D deep convolutional neural network, known as the Parkinsonism Differential Diagnosis Network (PDD-Net), to automatically identify imaging-related indices that could support the differential diagnosis of parkinsonian diseases. This model was used to examine parkinsonian PET imaging from two groups: more than 2,100 patients from China and 90 patients from Germany.
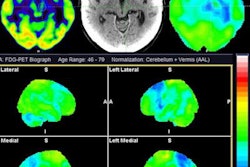
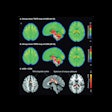
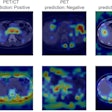
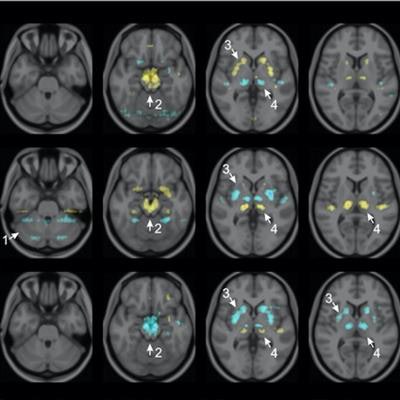
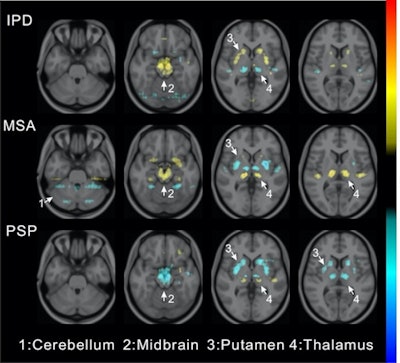 Visualization of average saliency maps of patients with idiopathic Parkinson's disease (IPD), multiple system atrophy (MSA), and progressive supranuclear palsy (PSP) in the training cohort showing characteristic regions contributing to the deep metabolic imaging (DMI) indices. The color corresponds to the importance score indicating the contribution of a region for the generated DMI indices. The color directions (yellow and red vs. cyan and blue) represent different influences on the DMI indices (increased uptake value contributes to the increase or decrease of the probability of IPD, MSA, or PSP in the DMI indices). The arrows pointed to the most salient brain regions including 1: Cerebellum, 2: Midbrain, 3: Putamen, and 4: Thalamus. Image courtesy of the Journal of Nuclear Medicine.
Visualization of average saliency maps of patients with idiopathic Parkinson's disease (IPD), multiple system atrophy (MSA), and progressive supranuclear palsy (PSP) in the training cohort showing characteristic regions contributing to the deep metabolic imaging (DMI) indices. The color corresponds to the importance score indicating the contribution of a region for the generated DMI indices. The color directions (yellow and red vs. cyan and blue) represent different influences on the DMI indices (increased uptake value contributes to the increase or decrease of the probability of IPD, MSA, or PSP in the DMI indices). The arrows pointed to the most salient brain regions including 1: Cerebellum, 2: Midbrain, 3: Putamen, and 4: Thalamus. Image courtesy of the Journal of Nuclear Medicine.The deep metabolic imaging indices extracted from PDD-Net provided an early and accurate method for the differential diagnosis of parkinsonian syndromes, with high rates of sensitivity and specificity for Parkinson's disease, multiple system atrophy, and progressive supranuclear palsy, according to the findings.
"Deep-learning technology may help physicians maximize the utility of nuclear medicine imaging in the future," Wu and colleagues concluded.