MRI radiomics features can differentiate breast cancers based on human epidermal growth factor receptors (HERs), according to research published January 24 in the American Journal of Roentgenology.
Researchers led by Yuqin Peng from Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University in Guangzhou, Guangdong found that their nomogram based on MRI radiomics can distinguish HER2-zero from HER2-low or HER2-positives cancers, as well as HER2-low from HER2-positive cancers.
“MRI radiomics may help select patients for novel or traditional HER2-targeted therapies, particularly in those with ambiguous immunohistochemical results or limited access to fluorescence in situ hybridization,” Peng and colleagues wrote.
According to guidelines by the American Society of Clinical Oncology and American Pathological Society, clinicians should go by a three-tiered system to determine breast cancer HER2 expression: HER2-zero (not eligible for HER2-targeted therapy), HER2-low (novel HER2-targeted drugs), and HER2-positive (traditional HER2-targeted medications). These expressions exhibit differing biological characteristics that influence treatment strategy.
While traditionally, pathologic analysis of biopsy or surgery specimens is used to determine expression status, the researchers noted that such analysis is susceptible to interobserver variability. They added that this could also lead to ambiguous interpretations in tumors with heterogeneous HER2 expression.
Previous studies have explored the potential MRI radiomics models in identifying breast cancer HER2 expression status. However, the researchers pointed out a lack of data on radiomics models in further distinguishing HER2-low from HER2-positive tumors.
Peng and co-authors analyzed the performance of MRI radiomics models for the three-tiered classification of HER2 expression status in breast cancer patients. They included 592 women with an average age of 47 years in the retrospective study. The women underwent breast MRI between 2016 and 2022.
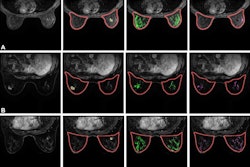
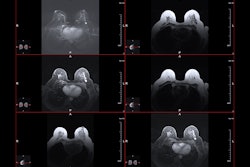
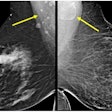
Radiologists assessed conventional MRI features for tumors and from there, manually segmented the tumors into multiparametric sequences to extract radiomic features. These included T2-weighed images, diffusion-weighted imaging (DWI), apparent diffusion coefficient (ADC), and delayed contrast-enhanced images.
The team used least-absolute shrinkage and selection operator analysis to develop two radiomics signatures. One differentiated HER2-zero from HER2-low or HER2-positive cancers (task 1), while the other distinguished HER2-low from HER2-positive cancers (task 2).
The researchers randomly assigned women from one hospital to discovery (task 1, n = 376; task 2, n = 335) or internal validation sets internal validation sets (task 1, n = 161, task 2, n = 143). Women at a second hospital meanwhile formed an external validation set (task 1, n = 55, task 2, n = 50).
The team reported that their radiomics method led to moderate-to-high performance in distinguishing breast cancers from different HER2 expression statuses.
| Performance of MRI radiomics on determining HER2 status |
||||||
|---|---|---|---|---|---|---|
| |
Task 1 | Task 2 | ||||
| Measure | Discovery | Internal validation | External validation | Discovery | Internal validation | External validation |
| Area under the curve (AUC) | 0.89 | 0.86 | 0.78 | 0.77 | 0.75 | 0.77 |
| Sensitivity | 99.4% | 98.6% | 100% | 93.8% | 92.9% | 97% |
| Specificity | 69% | 76.5% | 0% | 32.3% | 6.8% | 29.4% |
The researchers explained that no nomogram was created for task 1, since they found no significant ties between clinicopathologic or conventional MRI features and HER2 status that was independent of the MRI radiomics signature.
However, they found that for task 2, a nomogram including MRI radiomics signature and three pathologic features (histologic grade III, high Ki-67, positive progesterone receptor status) showed independent associations with HER2-low expression. This included AUC values of 0.87 (discovery), 0.83 (internal validation), and 0.8 (external validation).
The study authors highlighted that while MRI radiomics is not sufficient in being a first-line test in this area, it could aid clinicians in assessing HER2 status in breast cancer patients who have ambiguous expression status through other pathologic testing methods.
The authors also called for future studies to be multicentered, to include data from diverse patient populations for generalizability purposes, and to use computer-assisted segmentation methods for better standardization.
The study can be found in its entirety here.