Detecting changes in follow-up imaging studies can be difficult due to technical factors as well as human error. A software algorithm developed by a Canadian research team may be able to provide radiologists with some assistance.
A group led by Varvara Nika from the department of mathematics and statistics at York University in Toronto has developed a software algorithm that automatically performs image registration and calculates any changes between subsequent brain MRI studies. Experimental results with both simulated and real MRI scans show that the software, called EigenBlock Change Detection (EigenBlockCD), outperforms previous methods for detecting changes, according to the researchers.
"It detects clinical changes while ignoring the changes due to patient position and other acquisition artifacts," Nika said.
She presented the findings during a session at the SPIE Medical Imaging 2014 meeting in San Diego.
The challenge of change
It can be difficult to identify what has meaningfully changed in follow-up medical imaging studies. For example, differences between images could be due to acquisition-related factors such as radiofrequency inhomogeneities, registration changes from patient position, a different selection of acquisition pulse sequences, and hardware and software changes, Nika said.
In addition, radiologists might miss a change due to factors such as mistakenly classifying disease-related changes as acquisition-related changes, information overload, satisfaction of search, change in an unpredicted location, or a new change occurring in a complex lesion, according to the researchers.
While automatic methods for identifying changes in serial MR images taken at different times are of great interest to radiologists, most existing change-detection methods in medical imaging -- and for brain images, in particular -- include many preprocessing steps and rely mostly on statistical analysis of MRI scans.
"Although most methods utilize registration software, tissue classification remains a difficult and overwhelming task," Nika said.
As a result, the researchers sought to develop an automated algorithm that could detect subtle and disease-related changes on images. They also wanted to reduce the amount of data presented to the radiologist, as well as show the changes in a way that matches the human eye and makes them easy to read, see, and understand, she said.
Following up on previous work using dictionary learning techniques to automatically detect changes in MR images, Nika and colleagues Dr. Paul Babyn, from the University of Saskatchewan's department of medical imaging, and Hongmei Zhu, PhD, from York University's department of mathematics and statistics, simplified and improved their prior algorithm to develop EigenBlockCD.
Automatic change detection
EigenBlockCD uses principal component analysis to eliminate redundancy and increase computational efficiency, according to the researchers. No preprocessing steps are required, and the algorithm uses blocks of pixels from one scan as training samples to detect changes in the follow-up study.
"The main idea of EigenBlockCD is to perform a local image registration for identifying important structural changes, such as appearance, disappearance, growth, or shrinkage of a lesion, while automatically rejecting unimportant changes due to spatial position of patients, measurement noise, and common imaging acquisition artifacts," Nika told AuntMinnie.com.
In the first step, EigenBlockCD performs an initial global alignment of the images using fixed large radii of blocks of interest. Next, 2D images are aligned by performing a linear transformation of one MR image with its follow-up image via the use of two translation parameters and one rotation parameter. The algorithm then determines any significant changes between the two images by using fixed small blocks of interest.
"The EigenBlockCD algorithm divides the test image into many overlapping blocks," she said. "We assume that voxels from one image will appear again in the follow-up image either at the same location or within a close neighborhood from which the training blocks are learned."
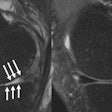
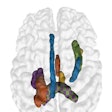
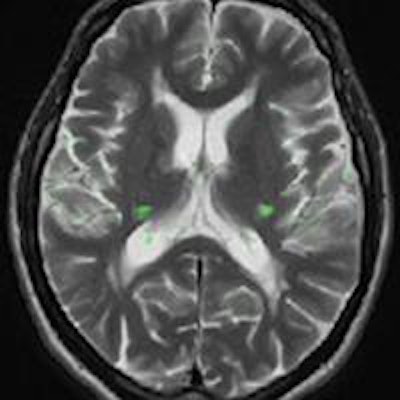
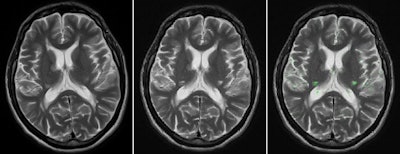 Brain MRI on same patient from 2011 (left) and 2013 (center). Overlaid changes (green) calculated by the EigenBlockCD algorithm (right). All images courtesy of Varvara Nika.
Brain MRI on same patient from 2011 (left) and 2013 (center). Overlaid changes (green) calculated by the EigenBlockCD algorithm (right). All images courtesy of Varvara Nika.The researchers compared the performance of EigenBlockCD with a simple differencing technique for computing changes between a ground truth image and a changed image. They then repeated the experiment.
In 48 experiments, EigenBlockCD yielded a mean 99.9% correct classification, 75.6% sensitivity (probability that the algorithm detected true changes), and 100% specificity (probability that the algorithm detected nonchanges). In contrast, the simple differencing method achieved 36% correct classification, 32.7% sensitivity, and 100% specificity.
"The EigenBlockCD algorithm achieves promising results," Nika said.
Next, the researchers plan to extend the algorithm to 3D volumetric data. They are also working on extending the algorithm to multispectral dictionary learning, she said.