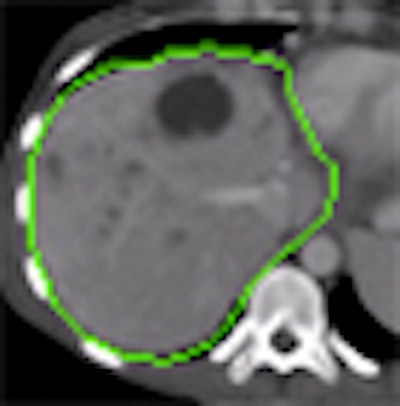
BERLIN - Liver segmentation is a critical but complex task that has challenged a generation of developers. The sheer variety of possible shapes and structures found in the organ, as well as the effects of contrast agents, tumors, fat, and even cirrhosis on CT imaging, can foil automated and semiautomated segmentation schemes alike. On the other hand, manual segmentation is too time-consuming for practical clinical use.
Researchers from medical image computing firm Fraunhofer Mevis in Bremen, Germany, believe they have found a way to preserve the best aspects of automated and manual segmentation, enabling users to easily correct mistakes in manual segmentation.
"Automatic algorithms do not produce correct results in all cases, but most parts of automatic segmentation are typically correct," said research scientist Frank Heckel at last week's Computer Assisted Radiology and Surgery (CARS) conference.
"One solution to the segmentation problem can be an automatic segmentation followed by manual correction," he said. "The correction method has to be local, fast, ideally three-dimensional, based on intuitive user interaction, and, last but not least, it would be nice if the algorithm was independent of the actual segmentation algorithm."
The firm has developed a liver segmentation algorithm based on an earlier tumor segmentation tool developed by the group (Proceedings of SPIE Medical Imaging, 2009: Image Processing, Vol. 7259). The manual correction component of the scheme is based on a method previously developed to correct segmentation in lung nodules, liver metastases, and lymph nodes.
Modifications were needed in the original algorithms due to the many differences between livers and tumors. Livers are larger and less compact, their surface is typically less smooth than that of a tumor, and the organ has a wide variety of structures surrounding it, Heckel said.
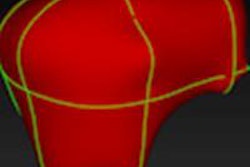
To correct a mistake in segmentation, the user simply draws in the correction as a 2D contour that defines the actual border of the object in one slice, Heckel said.
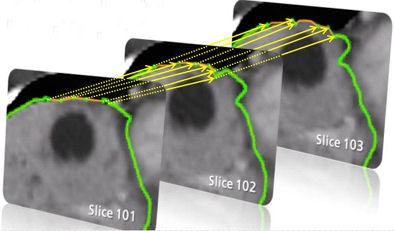
An extrapolation process is performed using a live-wire segmentation algorithm, which automatically moves the user-drawn contour to adjacent slices. The algorithm accomplishes this by moving equidistant seed points on the contour to the adjacent slice using a 2D block-matching algorithm.
 |
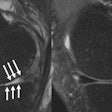
| Step one (above) requires the user to simply draw the actual border of the liver in one slice. The extrapolation of this user contour extracts equidistant seed points, connects seed points using a live-wire segmentation algorithm, and replaces part of the initial contour. Below, the user contour is extrapolated to neighboring slices for 3D correction. A search for seed points is performed in a given search area using a block matching algorithm. The seed points are connected using the live-wire algorithm, and this process continues until termination criteria are met. |
 |
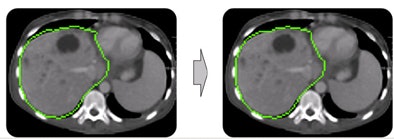 |
| In step two, morphological postprocessing ensures a coherent, smooth mass. The opening process removes small artifacts, connected component analysis removes separated parts, and closing eliminates small gaps. |
After these steps, the algorithm goes back and compares its modifications to the correction of the tumor segmentation that the user generated.
Performance of the algorithm has been improved by using multicore processor capabilities, and by adding methods to compensate for the large size of the liver, Heckel said.
Additional improvements "consist of parameter optimizations, producing better and more robust results in all test cases," Heckel said. "The distance between the seed points had to be adapted a little bit, and the size of the reference block for the block imaging algorithm had to be modified to locate the seed points more robustly."
The live-wire segmentation function was also modified with the addition of new termination conditions that tell the algorithm when to stop locating seed points on new slices, he said.
Testing
The modified algorithm was evaluated by four radiologists and one technical expert on 51 liver segmentations that were initially rated as insufficient (n = 24) or acceptable (n = 27), Heckel said.
Using a predefined upper time limit of five minutes for the manual correction, 82% of the segmentations were improved, and 88% of the "insufficient" segmentations were improved to "good" or "excellent" with use of the algorithm, Heckel said. Similarly, 78% of the "acceptable" segmentations were improved to "good" or "excellent."
"Our method can effectively be used for the correction of liver segmentations, and our algorithm is local, it's fast, and it generates 3D corrections based on an intuitive 2D user interaction," Heckel said.
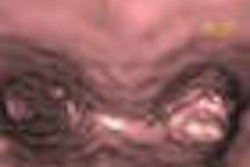
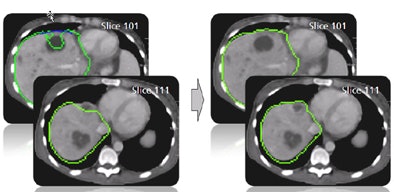 |
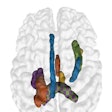
| The results demonstrate correction of the segmentation in 3D in the user-selected 2D slice, as well as automatic correction in the adjacent slices. |
The algorithm is independent of the initial segmentation algorithm and can be used for a wide variety of applications, he said. It is not limited to raw image data, but can be applied to multiplanar reformats, or sagittal or coronal views, he said. And a user who makes a mistake in drawing the correction can simply use the "undo" function to reverse the segmentation and start over.
"Our algorithm is a general approach that can be used for other objects and other modalities as well, as long as the objects to be segmented are visually separated from the background," he said. "The object needs to be coherent and compact."
Future plans include further improvements to the live-wire function, new "on-the-fly training" strategies that allow the algorithm to learn from experience, more-robust features, and additional performance improvements, he said. Finally, the group is looking at adapting the algorithm to other organs and modalities.
By Eric Barnes
AuntMinnie.com staff writer
July 1, 2009
Related Reading
Automated CT volumetry of lymph nodes gauges treatment response, March 27, 2009
Nodule measurement streamlined with space-scale analysis, November 25, 2008
Novel CAD distinguishes liver lesions on ultrasound, September 29, 2008
Measuring lung nodule growth beyond variability, November 5, 2007
Radiologists no match for nodule measurement software, August 31, 2007
Copyright © 2009 AuntMinnie.com